- The ChemDIS-Mixture function can only be accessed by selecting 'Mixture' link shown as the following figure.

- By the default setting, there are four fields required for analysis the overlapped effects of mixtures including two chemical names, score and database version as the following figure. Additional chemicals can be entered by clicking the [+] icon. Currently, ChemDIS support the analysis of maximum 4 chemicals. After entering characters in the 'Name' field, the autocomplete function will provide chemicals matching the keywords. Please note that the keyword is case-insensitive. Only matching chemicals are available in the ChemDIS system.
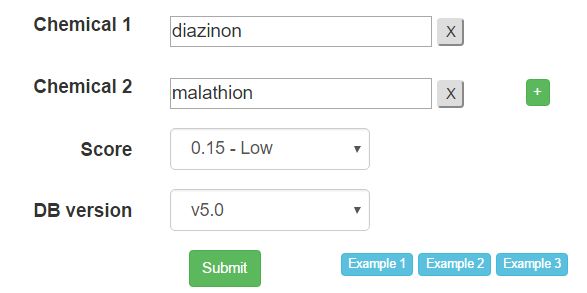
- There are three options for the 'Score'. the score define what proteins should be utilized for subsequent analysis. Default is to extract proteins >= 0.15. Please refer to the sction 'What is the score' for detail.
- There are two options for the 'DB version'. It decides which version of STITCH database (5.0 or 4.0) should be applied for analysis. Default is to use STITCH v5.0 for analysis.
Step.3) Click 'Submit Query'.
Step.4) The analysis result will be represented as Venn diagrams showing the overlapped effects of GO, pathway and DO terms as the following figure. The numbers are clickable for detail information.
- For example, there are 27 DO terms unique to diazinon and 374 terms unique to malathion, respectively. A total of 99 terms were the overlapped effects. In addition, 312 terms representing the overall effects were obtained from the analysis of all union proteins interacting the two chemicals.
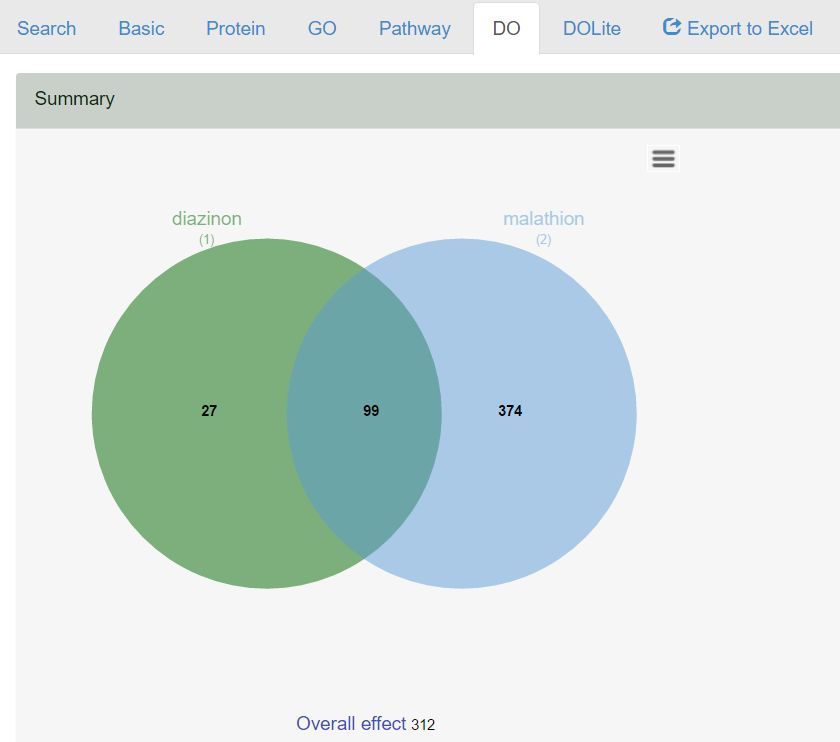
- For example, there are 27 DO terms unique to diazinon and 374 terms unique to malathion, respectively. A total of 99 terms were the overlapped effects. In addition, 312 terms representing the overall effects were obtained from the analysis of all union proteins interacting the two chemicals.
By clicking the link of the overlapped 99 terms, the detailed information will be shown in the bottom of the page. The Venn diagram will be collapsed that can be reopened by clicking the link [Click to expand].
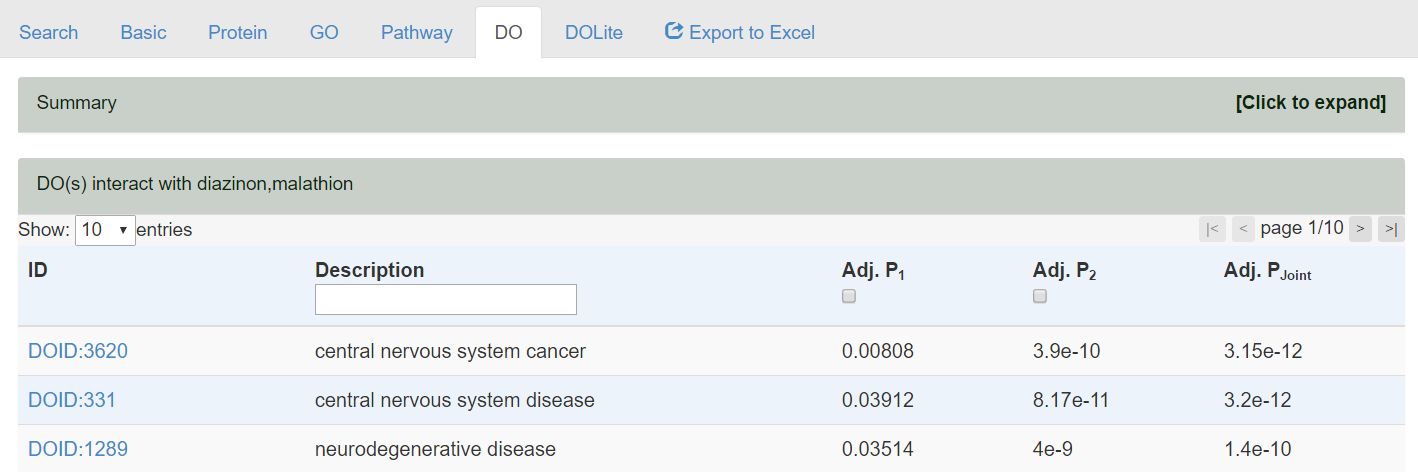
- The small check box in the header row is clickable for displaying the detailed analysis results for every cehcmials as the following figure. For the interpretation of the values, please refer to the corresponding section What are the columns in the result tables
- There will be individual p-values for every chemicals (Adj. P1, Adj. P2) and a joint p-value of Adj. Pjoint calculated by multiply Adj. P1 and Adj. P2.