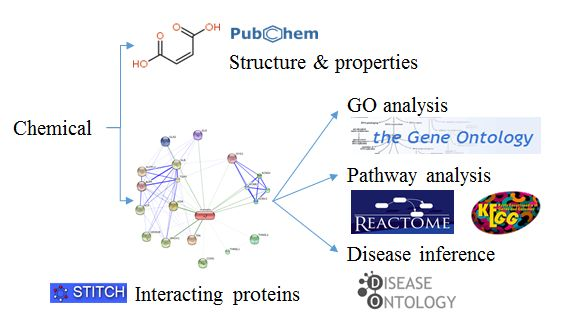
ChemDIS integrated several databases and packages for analyzing enriched terms. The analysis pipeline is listed as follows.
- The basic information of chemicals extracted from PubChem will be shown
- Given a chemical, the interacting proteins with scores larger than or equal to the user-selected threshold will firstly be extracted from STITCH database
- The enriched analysis of Gene Ontology (GO) terms, KEGG and reactome pathways, and Disease Ontology (DO) and DOLite disease terms will be conducted based on a hypergeometric test.
- Finally, the adjusted p-values will be calculated based on Benjamini-Hochberg multiple test correction.